A first for CRISPR: Cutting genes in blood stem cells
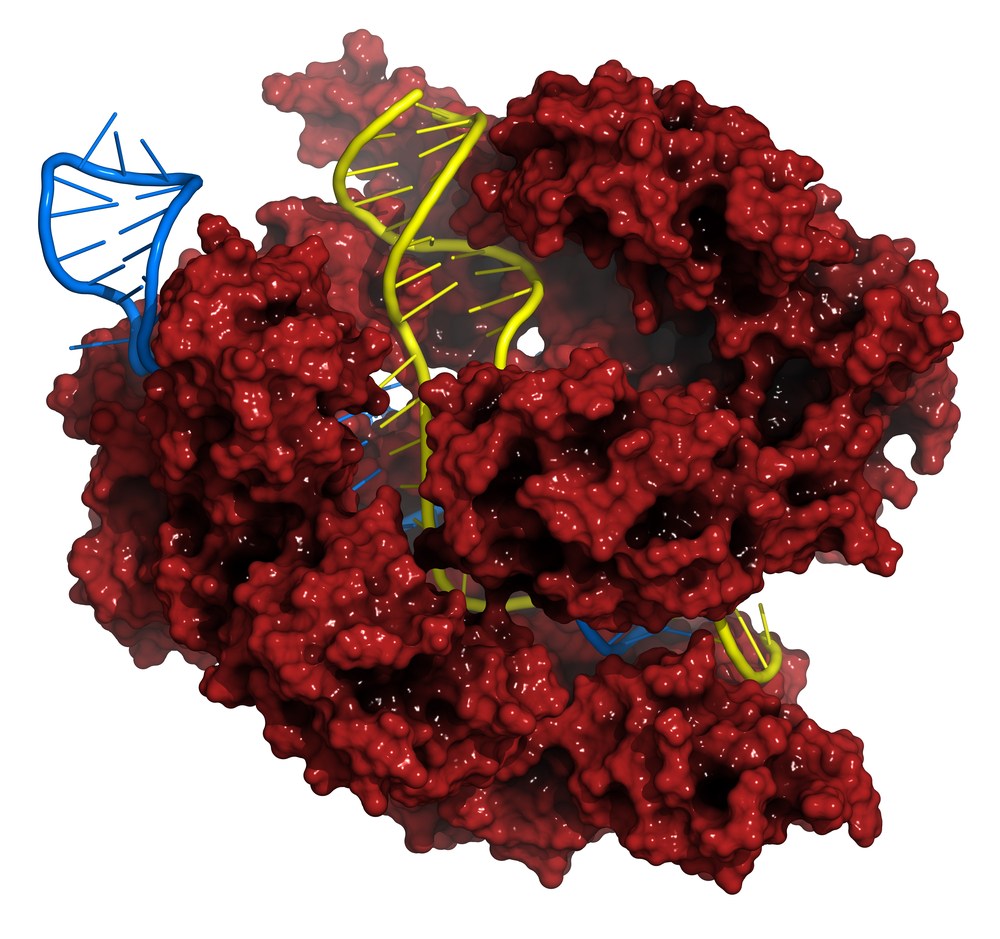
CRISPR — a gene editing technology that lets researchers make precise mutations, deletions and even replacements in genomic DNA — is all the rage among genomic researchers right now. First discovered as a kind of genomic immune memory in bacteria, labs around the world are trying to leverage the technology for diseases ranging from malaria to sickle cell disease to Duchenne muscular dystrophy.
In a paper published yesterday in Cell Stem Cell, a team led by Derrick Rossi, PhD, of Boston Children’s Hospital, and Chad Cowan, PhD, of Massachusetts General Hospital, report a first for CRISPR: efficiently and precisely editing clinically relevant genes out of cells collected directly from people. Specifically, they applied CRISPR to human hematopoietic stem and progenitor cells (HSPCs) and T-cells.
“CRISPR has been used a lot for almost two years, and report after report note high efficacy in various cell lines. Nobody had yet reported on the efficacy or utility of CRISPR in primary blood stem cells,” says Rossi, whose lab is in the hospital’s Program in Cellular and Molecular Medicine. “But most researchers would agree that blood will be the first tissue targeted for gene editing-based therapies. You can take blood or stem cells out of a patient, edit them and transplant them back.”
The study also gave the team an opportunity to see just how accurate CRISPR’s cuts are. Their conclusion: It may be closer to being clinic-ready than we thought.
Targeting blood cancers and HIV
With an eye toward clinical use, Cowan and Rossi, who collaborate in the Harvard Stem Cell Institute, initially targeted a gene called beta 2 microglobin (B2M) in T-cells, and another gene called CCR5 — the co-receptor for the human immunodeficiency virus (HIV) — in HSPCs.
“The idea with editing out B2M was to create a cell that can’t be recognized by the immune system,” Rossi explains, “and which could potentially be used clinically for things like CAR T-cell therapy [in cancer].”

The pair targeted CCR5 for a couple of reasons. “It’s been done before with other technologies, so we had something to compare to,” says Rossi. “But it also plays to the strength of CRISPR, which is gene knockout.”
The other work Rossi refers to is by the biotechnology company Sangamo BioSciences. Inspired by the “Berlin patient” — an HIV patient who was cured of the virus after receiving a stem cell transplant from a donor whose T-cells carried a crippled version of CCR5 — Sangamo is trying to knock CCR5 out of patients’ cells using a competing gene editing technology called zinc finger nucleases (ZFNs). The company’s work is already in Phase 2 clinical trials.
Cuts, with precision
In their Cell Stem Cell paper, Rossi and Cowan showed they could edit B2M out of T-cells and CCR5 out of HSPCs efficiently, predictably and precisely. They further showed that the edited HSPCs could go on to produce the normal portfolio of blood and immune cells.
The pair also addressed concerns about just how exact CRISPR is.
“There have been a couple of papers suggesting that CRISPR has high ‘off-target’ activity,” Rossi explains. “We decided to test its accuracy under six different experimental conditions.”
The team performed deep sequencing on sites in the genome that could possibly confuse CRISPR when trying to edit CCR5 out of HSPCs. Making an average of 3,400 sequencing passes (compared to the 50 usually used for whole genome sequencing) they found that the system’s risk of making aberrant cuts was effectively zero.
“That’s preclinical evidence in a relevant cell type that version 1.0 of CRISPR, when used judiciously, is ready for patients,” Rossi says.
Rossi says the team’s data open up the possibility of using CRISPR to go after enhancers—DNA switches that help turn genes on and off—for genes like BCL11a, a hot target in sickle cell disease. “Say you know there’s a 5,000-base-pair region that controls the expression of BCL11a. You could cut the whole enhancer out without touching the gene at all.”
Read more about Rossi and Cowan’s study in the Harvard Gazette.
Related Posts :
-

Tough cookie: Steroid therapy helps Alessandra thrive with Diamond-Blackfan anemia
Two-year-old Alessandra is many things. She’s sweet, happy, curious, and, according to her parents, Ralph and Irma, a budding ...
-

A new druggable cancer target: RNA-binding proteins on the cell surface
In 2021, research led by Ryan Flynn, MD, PhD, and his mentor, Nobel laureate Carolyn Bertozzi, PhD, opened a new chapter ...
-

Unique data revealed just when Mickey’s heart doctors could operate
When Mikolaj “Mickey” Karski’s family traveled from Poland to Boston to get him heart care, they weren’t thinking ...
-

Forecasting the future for childhood cancer survivors
Children are much more likely to survive cancer today than 50 years ago. Unfortunately, as adults, many of them develop cardiovascular ...