Hidden part of flu virus yields hope for better vaccines
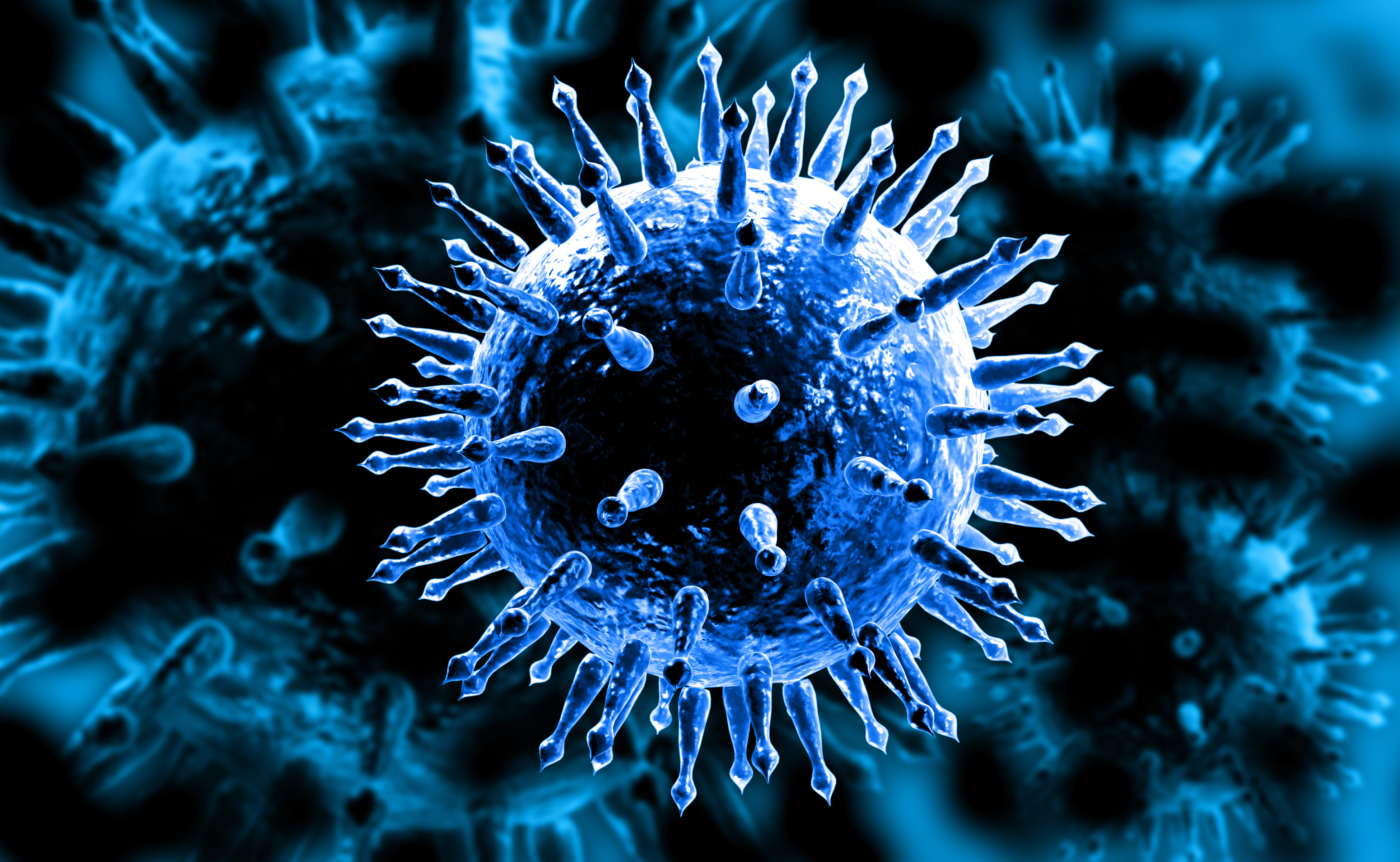
The annual effort to create a flu vaccine is like a high-stakes game of Whack-a-mole. Even if public health officials develop a vaccine that offers wide protection, the ever-changing influenza virus pops up again the next year in a new guise, often different enough to thwart the body’s defenses. And while most people recover, influenza typically kills more than ten thousand people a year in the U.S. — and hundreds of thousands around the world.
Recently, scientists have been attempting to end this vicious cycle by creating what the National Institutes of Health calls a “universal” flu vaccine — one that offers protection against many strains of the virus, year after year. One strategy has been to target two parts of the virus that are so essential to its survival that they don’t change as the virus evolves. In this week’s issue of Cell, scientists identify a third such potential target in a previously unknown location.
“This could be a step toward a next-generation flu vaccine that’s substantially more effective than the ones we have today,” says Stephen Harrison, PhD, chief of the Division of Molecular Medicine at Boston Children’s Hospital and a senior co-author of the paper along with Garnett Kelsoe, D.Sc., of Duke University.
Hijacking the machinery of human cells

known as a flail.
IMAGE: ADOBE STOCK
The influenza virus resembles the kind of spike-studded iron ball once used as medieval weapon. Most of the spikes on its surface are made of a protein called hemagglutinin, which, up close, looks a bit like a three-headed mushroom.
Hemagglutinin serves two critical functions: It allows the flu virus to stick to the surface of cells in the linings of the human nose, throat and lungs. Then it penetrates those membranes and inserts the viral genes, which commandeer the cell’s genetic machinery. “The virus hijacks the cell and tells it: ‘Make lots more of me and none of you,’” Harrison says.
A single human cell can make thousands of copies of the virus, which are then released to infect more cells.
Arms race vs. shape-shifter
In response to this attack, the immune system’s B cells make antibodies that recognize particular parts, or “antigens,” of the virus, especially hemagglutinin. By latching onto those antigens, the antibodies prevent the viruses from entering any more cells, eventually bringing the infection to an end.

But the flu virus is, literally, a shape-shifter. Random mutations in its genes continually result in small changes to the virus’s surface. By chance, some of those changes interfere with the “fit” of antibodies formed during previous exposures to the flu. Thanks to natural selection — “survival of the fittest” — these new forms of the virus multiply and become the dominant strain in the next flu season.
“The virus evolves,” Harrison says. “It changes the characteristics of its surface to weaken the binding of those antibodies and hence escape immunity. This is the virus’s side of the arms race with our immune system.”
Seeking out the virus’s vulnerable spots
Looking for a way to end this arms race, scientists have been trying to identify parts of hemagglutinin that can’t easily mutate because they’re critical to the virus’s survival. Two such parts are well known: the stem of the mushroom and the “receptor binding sites” — the points on each of the mushroom’s three caps that stick to human cells.
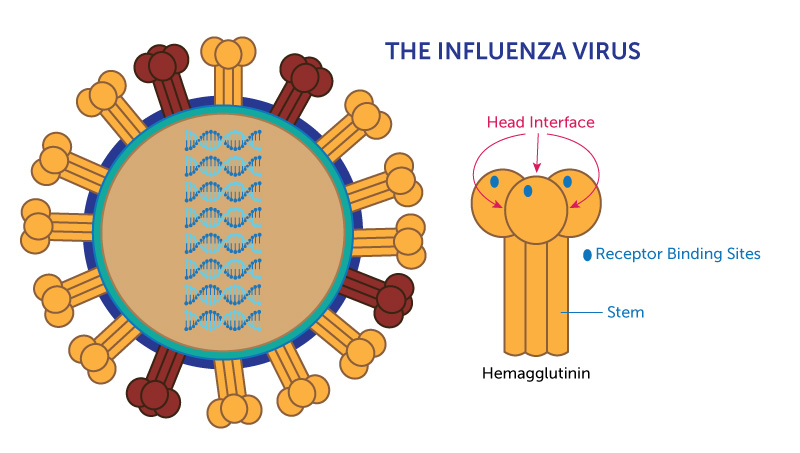
Because these parts change little from year to year, the hope is that vaccines targeting these “conserved” antigens might offer broader and longer-lasting protection from flu. The National Institutes of Health recently began a clinical trial of a vaccine designed to stimulate antibodies to the stem of the hemagglutinin protein. Similar trials are under way around the world.
However, the immune system responds more strongly to the head of the hemagglutinin protein than to the stem. And last year, some of the researchers involved in the Cell paper made an intriguing discovery: When studying B cells from people recently given vaccines, they found that some B-cell receptors recognized hemagglutinin in multiple strains of the flu virus. Three-quarters of those receptors recognized sites on the head, not the stem, of hemagglutinin. But most of these B cells did not recognize the receptor binding site, the only known conserved site on the head. These results made the researchers wonder: Might there be other, undiscovered stable regions of the hemagglutinin head shared by multiple forms of the virus? The Duke-Boston Children’s team launched a search to find out.
Revealing a hidden flu target
They collected B cell samples from four adults before and after they were given flu shots. Then they screened those samples for antibodies produced in response to two types of flu. After eliminating the antibodies to the known conserved parts of the hemagglutinin head and stem, they used X-ray crystallography to help visualize where the remaining antibodies were binding. They found that some of them bound to the points where hemagglutinin’s three mushroom caps touch, a region they call the head interface.
The finding was surprising, Harrison says, because these points are normally physically hidden from the immune system. Indeed, head-interface antibodies offered no protection against the flu virus to cells in a lab dish.
But when some of Harrison’s co-authors injected the virus into live mice, they found that the head-interface antibodies offered “robust” protection. That’s because “all proteins tend to breathe open and shut a bit,” Harrison says. This molecular flexing gives enough time for the immune system to form antibodies — and, later, for those antibodies to bind to these normally hidden spots in the virus’s surface.
It’s easier for the immune system to access the head interface when hemagglutinin is present on the surface of an infected cell. So these antibodies appear to provide immunity not by preventing viruses from entering cells but by recruiting the immune system’s natural killer cells to dispatch infected cells before the viruses can escape to do more damage.
Game plan for better vaccines
Harrison does not believe this discovery is a “home run” in the search for a universal flu vaccine. “But we don’t necessarily need a home run,” he says, “because you can win a ballgame with a lot of singles. My own view is that we will get better flu vaccines, ones that give immunity that’s less sensitive to viral variation. But I think we’re going to get there by a bunch of singles.”

Harrison sees a parallel in today’s treatments for HIV. Many patients are kept alive not by a single drug but with a cocktail of three drugs given at the same time. The HIV virus might be able to evolve resistance to one drug, even two, but the chances of becoming resistant to all three at once are very small.
Similarly, Harrison says, if future vaccines can be designed to stimulate antibodies to all three stable regions on the flu virus, they might offer longer lasting protection against new strains, including wholly new ones with the potential to cause a global pandemic.
The discovery, to use Harrison’s baseball analogy, is like a line drive up the middle — not a game-winner, but one hit in a rally that might give us a lead in the never-ending struggle against a wily opponent.
Akiko Watanabe, PhD, a researcher in the Duke University Department of Immunology, and Kevin R. McCarthy, PhD, a postdoc in the Laboratory of Molecular Medicine at Boston Children’s Hospital, were first co-authors on the paper. Kelsoe, of the Duke University Vaccine Institute, and Harrison were senior co-authors. For a complete list of co-authors, see Cell. This work was supported by grants from the National Institutes of Health and the National Institute of General Medical Sciences.
Related Posts :
-

Creating the next generation of mRNA vaccines
During the COVID-19 pandemic, mRNA vaccines came to the rescue, developed in record time and saving lives worldwide. Researchers in ...
-

Tracking influenza in its first battleground: The nose
The answer to curbing influenza could be right under our noses — or, more accurately, inside them. New research maps happenings ...
-

From ‘hit to vial’: Discovery and optimization of a promising vaccine adjuvant
Many vaccines are only partially effective, have waning efficacy, or do not work well in the very young or the ...
-

Boosting vaccines for the elderly with ‘hyperactivators’
As we age our immune systems start to flag, leaving us more susceptible to cancer and infections — and less responsive ...